Microbes in gastrointestinal health and disease
- PMID: 19026645
- PMCID: PMC2892787
- DOI: 10.1053/j.gastro.2008.10.080
Microbes in gastrointestinal health and disease
Abstract
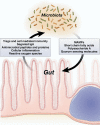
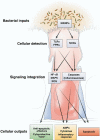
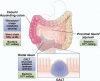
Most, if not all, animals coexist with a complement of prokaryotic symbionts that confer a variety of physiologic benefits. In humans, the interaction between animal and bacterial cells is especially important in the gastrointestinal tract. Technical and conceptual advances have enabled rapid progress in characterizing the taxonomic composition, metabolic capacity, and immunomodulatory activity of the human gut microbiota, allowing us to establish its role in human health and disease. The human host coevolved with a normal microbiota over millennia and developed, deployed, and optimized complex immune mechanisms that monitor and control this microbial ecosystem. These cellular mechanisms have homeostatic roles beyond the traditional concept of defense against potential pathogens, suggesting these pathways contribute directly to the well-being of the gut. During their coevolution, the bacterial microbiota has established multiple mechanisms to influence the eukaryotic host, generally in a beneficial fashion, and maintain their stable niche. The prokaryotic genomes of the human microbiota encode a spectrum of metabolic capabilities beyond that of the host genome, making the microbiota an integral component of human physiology. Gaining a fuller understanding of both partners in the normal gut-microbiota interaction may shed light on how the relationship can go awry and contribute to a spectrum of immune, inflammatory, and metabolic disorders and may reveal mechanisms by which this relationship could be manipulated toward therapeutic ends.
Figures
Similar articles
-
Composition and function of the human-associated microbiota.Nutr Rev. 2009 Nov;67 Suppl 2:S164-71. doi: 10.1111/j.1753-4887.2009.00237.x. Nutr Rev. 2009. PMID: 19906220 Review.
-
Microbiota-stimulated immune mechanisms to maintain gut homeostasis.Curr Opin Immunol. 2010 Aug;22(4):455-60. doi: 10.1016/j.coi.2010.06.008. Epub 2010 Jul 23. Curr Opin Immunol. 2010. PMID: 20656465 Review.
-
Recent advances in the role of probiotics in human inflammation and gut health.J Agric Food Chem. 2012 Aug 29;60(34):8249-56. doi: 10.1021/jf301903t. Epub 2012 Aug 16. J Agric Food Chem. 2012. PMID: 22897745 Review.
-
Prebiotic effects: metabolic and health benefits.Br J Nutr. 2010 Aug;104 Suppl 2:S1-63. doi: 10.1017/S0007114510003363. Br J Nutr. 2010. PMID: 20920376 Review.
-
The interplay between the gut microbiota and the immune system.Gut Microbes. 2014 May-Jun;5(3):411-8. doi: 10.4161/gmic.29330. Epub 2014 Jun 12. Gut Microbes. 2014. PMID: 24922519 Free PMC article. Review.
Cited by
-
Role of secretory IgA in the mucosal sensing of commensal bacteria.Gut Microbes. 2014;5(6):688-95. doi: 10.4161/19490976.2014.983763. Gut Microbes. 2014. PMID: 25536286 Free PMC article. Review.
-
Shotgun metagenomic sequencing analysis of ocular surface microbiome in Singapore residents with mild dry eye.Front Med (Lausanne). 2022 Nov 10;9:1034131. doi: 10.3389/fmed.2022.1034131. eCollection 2022. Front Med (Lausanne). 2022. PMID: 36438051 Free PMC article.
-
Microbiomes other than the gut: inflammaging and age-related diseases.Semin Immunopathol. 2020 Oct;42(5):589-605. doi: 10.1007/s00281-020-00814-z. Epub 2020 Sep 30. Semin Immunopathol. 2020. PMID: 32997224 Free PMC article. Review.
-
Restoring specific lactobacilli levels decreases inflammation and muscle atrophy markers in an acute leukemia mouse model.PLoS One. 2012;7(6):e37971. doi: 10.1371/journal.pone.0037971. Epub 2012 Jun 27. PLoS One. 2012. PMID: 22761662 Free PMC article.
-
A novel strain of Bacteroides fragilis enhances phagocytosis and polarises M1 macrophages.Sci Rep. 2016 Jul 6;6:29401. doi: 10.1038/srep29401. Sci Rep. 2016. PMID: 27381366 Free PMC article.
References
-
- McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. - PubMed
-
- McFall-Ngai MJ. Identifying “prime suspects”: symbioses and the evolution of multicellularity. Comp Biochem Physiol B Biochem Mol Biol. 2001;129:711–723. - PubMed
-
- McFall-Ngai MJ. Unseen forces: the influence of bacteria on animal development. Dev Biol. 2002;242:1–14. - PubMed
-
- Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. - PubMed
-
- Hickman C. How have bacteria contributed to the evolution of multicellular animals? In: McFall-Ngai MJ, editor. The influence of cooperative bacteria on animal host biology. Cambridge University Press; New York: 2005. pp. 3–32.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

