The Ensembl Variant Effect Predictor
- PMID: 27268795
- PMCID: PMC4893825
- DOI: 10.1186/s13059-016-0974-4
The Ensembl Variant Effect Predictor
Abstract
The Ensembl Variant Effect Predictor is a powerful toolset for the analysis, annotation, and prioritization of genomic variants in coding and non-coding regions. It provides access to an extensive collection of genomic annotation, with a variety of interfaces to suit different requirements, and simple options for configuring and extending analysis. It is open source, free to use, and supports full reproducibility of results. The Ensembl Variant Effect Predictor can simplify and accelerate variant interpretation in a wide range of study designs.
Keywords: Genome; NGS; SNP; Variant annotation.
Figures
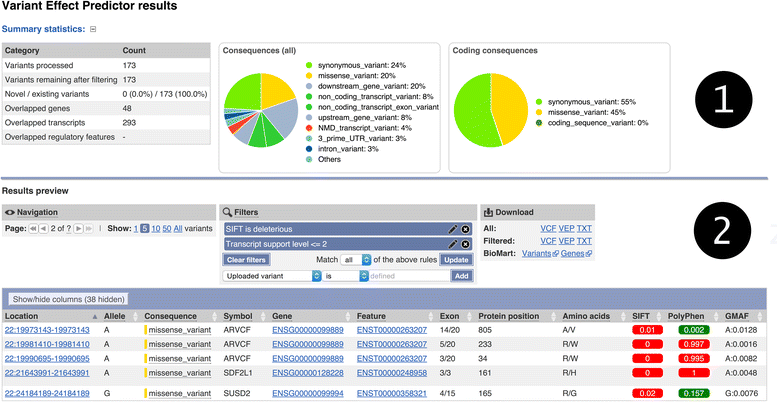

Similar articles
-
Ensembl 2021.Nucleic Acids Res. 2021 Jan 8;49(D1):D884-D891. doi: 10.1093/nar/gkaa942. Nucleic Acids Res. 2021. PMID: 33137190 Free PMC article.
-
Annotating and prioritizing genomic variants using the Ensembl Variant Effect Predictor-A tutorial.Hum Mutat. 2022 Aug;43(8):986-997. doi: 10.1002/humu.24298. Epub 2021 Dec 2. Hum Mutat. 2022. PMID: 34816521 Free PMC article.
-
Ensembl 2015.Nucleic Acids Res. 2015 Jan;43(Database issue):D662-9. doi: 10.1093/nar/gku1010. Epub 2014 Oct 28. Nucleic Acids Res. 2015. PMID: 25352552 Free PMC article.
-
In Silico Functional Annotation of Genomic Variation.Curr Protoc Hum Genet. 2016 Jan 1;88:6.15.1-6.15.17. doi: 10.1002/0471142905.hg0615s88. Curr Protoc Hum Genet. 2016. PMID: 26724722 Free PMC article. Review.
-
Genome information resources - developments at Ensembl.Trends Genet. 2004 Jun;20(6):268-72. doi: 10.1016/j.tig.2004.04.002. Trends Genet. 2004. PMID: 15145580 Review.
Cited by
-
Collateral sensitivity counteracts the evolution of antifungal drug resistance in Candida auris.Nat Microbiol. 2024 Nov;9(11):2954-2969. doi: 10.1038/s41564-024-01811-w. Epub 2024 Oct 29. Nat Microbiol. 2024. PMID: 39472696
-
Loss of TANGO1 Leads to Absence of Bone Mineralization.JBMR Plus. 2021 Jan 13;5(3):e10451. doi: 10.1002/jbm4.10451. eCollection 2021 Mar. JBMR Plus. 2021. PMID: 33778321 Free PMC article.
-
Variants at the ASIP locus contribute to coat color darkening in Nellore cattle.Genet Sel Evol. 2021 Apr 28;53(1):40. doi: 10.1186/s12711-021-00633-2. Genet Sel Evol. 2021. PMID: 33910501 Free PMC article.
-
Genomic Characteristics and Selection Signatures in Indigenous Chongming White Goat (Capra hircus).Front Genet. 2020 Aug 21;11:901. doi: 10.3389/fgene.2020.00901. eCollection 2020. Front Genet. 2020. PMID: 32973871 Free PMC article.
-
Introducing AI to the molecular tumor board: one direction toward the establishment of precision medicine using large-scale cancer clinical and biological information.Exp Hematol Oncol. 2022 Oct 31;11(1):82. doi: 10.1186/s40164-022-00333-7. Exp Hematol Oncol. 2022. PMID: 36316731 Free PMC article. Review.
References
-
- World Health Organisation. Non-communicable diseases: fact sheet. Jan 2015. http://www.who.int/mediacentre/factsheets/fs355/en/. Accessed 17 Mar 2016.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

