Identification of a foetal epigenetic compartment in adult human kidney
- PMID: 33783321
- PMCID: PMC8920245
- DOI: 10.1080/15592294.2021.1900027
Identification of a foetal epigenetic compartment in adult human kidney
Abstract
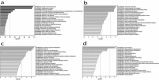
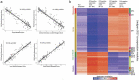
The mammalian kidney has extensive repair capacity; however, identifying adult renal stem cells has proven elusive. We applied an epigenetic marker of foetal cell origin (FCO) in diverse human tissues as a probe for developmental cell persistence, finding a 5.4-fold greater FCO proportion in kidney. Normal kidney FCO proportions averaged 49% with extensive interindividual variation. FCO proportions were significantly negatively correlated with immune-related gene expression and positively correlated with genes expressed in the renal medulla, including those involved in renal organogenesis (e.g., FGF2, PAX8, and HOXB7). FCO associated genes also mapped to medullary nephron segments in mouse and rat, suggesting evolutionary conservation of this cellular compartment. Renal cancer patients whose tumours contained non-zero FCO scores survived longer. The kidney appears unique in possessing substantial foetal epigenetic features. Further study of FCO-related gene methylation may elucidate regenerative regulatory programmes in tissues without apparent discrete stem cell compartments.
Keywords: DNA methylation; Kidney; epigenetics; foetal stem cells; stem cell niche.
Conflict of interest statement
J.K.W. and K.T.K. are cofounders of Cellintec.
Figures




Similar articles
-
Absence of an embryonic stem cell DNA methylation signature in human cancer.BMC Cancer. 2019 Jul 19;19(1):711. doi: 10.1186/s12885-019-5932-6. BMC Cancer. 2019. PMID: 31324166 Free PMC article.
-
Epigenetic features of testicular germ cell tumours in relation to epigenetic characteristics of foetal germ cells.Int J Dev Biol. 2013;57(2-4):309-17. doi: 10.1387/ijdb.130142ka. Int J Dev Biol. 2013. PMID: 23784842 Review.
-
DNA methylation analysis of Homeobox genes implicates HOXB7 hypomethylation as risk factor for neural tube defects.Epigenetics. 2015;10(1):92-101. doi: 10.1080/15592294.2014.998531. Epub 2015 Jan 29. Epigenetics. 2015. PMID: 25565354 Free PMC article.
-
Homeobox gene Rhox5 is regulated by epigenetic mechanisms in cancer and stem cells and promotes cancer growth.Mol Cancer. 2011 May 24;10:63. doi: 10.1186/1476-4598-10-63. Mol Cancer. 2011. PMID: 21609483 Free PMC article.
-
Investigation of epigenetics in kidney cell biology.Methods Cell Biol. 2019;153:255-278. doi: 10.1016/bs.mcb.2019.04.015. Epub 2019 Jun 13. Methods Cell Biol. 2019. PMID: 31395383 Free PMC article. Review.
Cited by
-
Recursive partitioning analysis for survival stratification and early imaging prediction of molecular biomarker in glioma patients.BMC Cancer. 2024 Jul 9;24(1):818. doi: 10.1186/s12885-024-12542-w. BMC Cancer. 2024. PMID: 38982347 Free PMC article.
References
-
- Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4(1–2):7–25. - PubMed
-
- Crane GM, Jeffery E, Morrison SJ. Adult haematopoietic stem cell niches. Nat Rev Immunol. 2017;17(9):573–590. - PubMed
-
- Tan DW, Barker N. Intestinal stem cells and their defining niche. Curr Top Dev Biol. 2014;107:77–107. - PubMed
-
- Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7(2):150–161. - PubMed
-
- Witzgall R, Brown D, Schwarz C, et al. Localization of proliferating cell nuclear antigen, vimentin, c-Fos, and clusterin in the postischemic kidney. Evidence for a heterogenous genetic response among nephron segments, and a large pool of mitotically active and dedifferentiated cells. J Clin Invest. 1994;93(5):2175–2188. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
